FUNCTIONAL PROTEOMICS – Neurodegenerative diseases
Functional proteomics
Proteins as the key to diagnostics
In the competence area of medical proteome analysis, one research focus is on the identification and validation of protein changes that occur in connection with neurodegenerative diseases. To this end, we are developing, among other things, highly sensitive mass spectrometry-based approaches for proteome and protein analysis. For prognostic and predictive diagnostics, we identify protein signatures that can potentially be used as disease-relevant biomarkers. We use spinal fluid and plasma to identify biomarkers. In addition, we focus on researching the underlying disease mechanisms. By means of specific tissue analyses, in which we use a combined laser microdissection-mass spectrometry approach, molecular processes are identified at the protein level, which can provide fundamental insights for diagnostic and therapeutic approaches.
Protein signatures for diagnostics
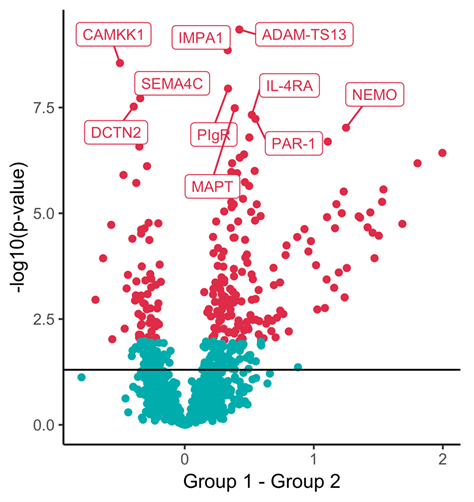
Neurodegenerative diseases, such as Alzheimer’s and Parkinson’s, are characterised by a progressive loss of nerve cells and the lack of adequate diagnosis and treatment. Many neurodegenerative diseases are described as proteinopathies and are characterised by the accumulation of disease-specific protein aggregates in the brain. Spinal fluid/ CSF, which is in direct contact with the central nervous system, plays a central role in diagnostics, because changes in the brain can lead to, among other things, abnormal protein abundance or altered protein modifications in the CSF. The affected proteins represent potential biomarkers that can be used for diagnostics. Known disease-associated proteins include amyloid-β peptides, tau or alpha-synuclein. We use high-resolution mass spectrometry to identify and quantify biomarkers. This method allows us to identify disease-specific protein signatures on a global scale in differential clinical studies (comparison of healthy versus diseased). We have already been able to find the first preliminary protein biomarker signatures in the cerebrospinal fluid for the early diagnosis of Parkinson’s disease. One challenge in identifying biomarkers is the respective extent of the change. The degree of disease-related abnormal protein abundance is very low at the beginning of a disease and may be almost impossible to measure due to the sensitivity and specificity of the analytical technique. For this, we are developing specific and highly sensitive targeted mass spectrometry-based approaches (e.g. Barkovits et al. J Proteome Res. 2018; Barkovits et al. Methods Mol Biol. 2019; Hörmann et al. Methods Mol Biol. 2019; Barkovits et al. Cells 2020; Barkovits et al. Mol Cell Proteomics 2020).
Uncovering neurodegenerative mechanisms
Neurodegenerative diseases primarily affect nerve cells of the human nervous system. Nerve cells are highly specialised cells that are responsible for information transmission and processing in the brain. Compared to other cell types, nerve cells basically cannot replace themselves, so dead nerve cells cannot be renewed by the body. In diseases of the nervous system, there is usually a loss of nerve cells in certain areas of the brain. In Parkinson’s disease, for example, nerve cells degenerate selectively in the substantia nigra pars compacta, while in Alzheimer’s disease there is a global degeneration of nerve cells in the hippocampus and cerebral cortex. Depending on the area affected, the loss of nerve cells leads to impaired movement or mental abilities and even death. To elucidate how affected tissue differs from healthy tissue and why different areas of the brain are affected in different neurodegenerative diseases, we use a laser microdissection-based approach. Laser microdissection allows us to isolate specific cellular as well as subcellular areas of the brain, allowing us to target the structures affected by the disease. By coupling laser microdissection with highly sensitive mass spectrometry, we can generate specific proteome profiles and thus obtain a global overview of processes that underlie normal/healthy as well as disease-related altered cellular processes at the protein level. We have already succeeded in identifying disease-specific changes in the proteome of individual neuron populations (Steinbach et al., in preparation) and of the pigment neuromelanin (Plum et al., 2016).