Method development
Proteomic methods for clinical questions
Proteome analyses using mass spectrometry
In addition to the identification of disease-relevant protein biomarkers for neurodegenerative diseases and cancer, the identification of underlying molecular and biochemical mechanisms is the focus of the research work of the competence area Medical Proteome Analysis. Especially for the identification of protein biomarkers, proteome analysis is a much-used approach. The proteome represents the totality of the expressed proteins of a cell, an organism or a body fluid under precisely defined conditions at a specific point in time. In contrast to the DNA in the genome, the proteome is very dynamic, as protein expression is influenced by many external factors such as metabolic state or environmental factors, and a specific protein pattern is expressed depending on these factors. In a cell, not all genes are switched on and translated into proteins at the same time. This results in specific proteome profiles for organisms but also for organs as well as cells and individual body fluids, which allow a targeted analysis known as proteomics. Key technology for proteomics is the combination of liquid chromatography and mass spectrometry, which can be used to identify several thousand proteins in a single complex sample in just a few hours. Proteomics research thus provides a global overview of the processes underlying healthy and diseased cellular processes at the protein level.
In the competence area of medical proteome analysis, we use state-of-the-art mass spectrometers for proteome analyses. Of particular note is the high-performance Orbitrap Fusion Lumos Tribrid mass spectrometer from Thermo Scientific, which was acquired as part of the Prodi research building and which we use for demanding applications.

Figure 1: A view of the workstation at the high-performance Orbitrap Fusion Lumos Tribrid mass spectrometer.
For clinical questions, proteome research offers us a global view of processes that correspond to healthy and diseased cellular processes at the protein level and thus offers the possibility to investigate the following aspects:
Protein identification
Here, all proteins in an examined sample are identified. Which proteins are formed under normal conditions in a particular cell type, tissue or body fluid?
Protein quantification
Here, an analysis of the (total) protein abundance in a particular cell type, tissue or body fluid is carried out. Which proteins are differentially expressed?
Post-translational modifications
Here we look for disease-relevant modifications to proteins. Does a protein carry a specific post-translational modification?
Protein localisation
Here we are looking for the localisation of a protein within the cell/tissue. Where is a protein formed and/or accumulates?
Functional proteomics
Here we identify the biological functions of specific individual proteins, protein classes or entire protein interaction networks. How does the function of a protein change depending on its quantity, modification, localisation or network?
Protein-protein interactions
Here we look at changes in a protein network depending on the disease state. Which proteins interact with each other (e.g. when and where)?
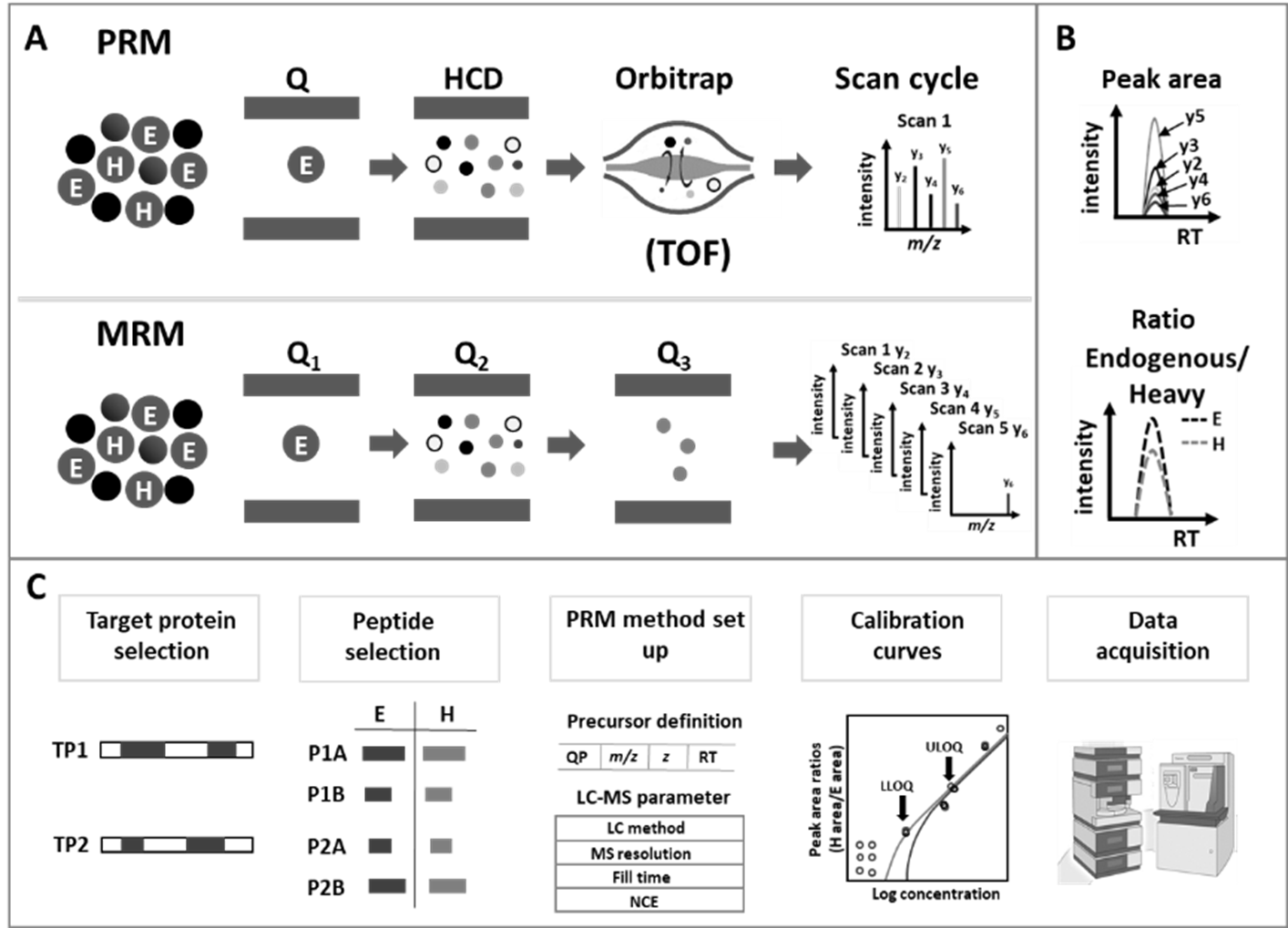
Figure 2: Principle of data acquisition, evaluation and workflow for targeted proteomics quantification.
Optimisation of biomarker detection
In order to enable timely but also specific clinical diagnostics and thus to be able to initiate preventive action options, there is a particular interest in the need for sensitive, specific and reliable methods for the detection of biomarkers. One of our research goals is to develop or optimise methods for already known protein biomarkers associated with the diseases (such as amyloid-β peptide, tau or alpha-synuclein for neurodegenerative disease or AHNAK2 for bladder cancer), with a focus on reproducible and robust analysis. For this purpose, we use a targeted proteomic approach that allows specific and sensitive detection.
Example: Detection of amyloid-β peptides
Amyloid-β peptides have been the focus of Alzheimer’s research for decades and have been approved as CSF biomarkers for Alzheimer’s diagnosis since 2011. However, the low concentration of the peptides poses a challenge for modern quantification techniques. Therefore, there is a great need for robust and sensitive amyloid-β quantification methods in body fluids such as CSF and plasma. In addition, there is increasing evidence that truncated forms of amyloid-β peptides play a central role in Alzheimer’s disease. In the competence area of medical proteome analysis, we optimise and develop mass spectrometry-based methods for the detection of amyloid-β peptides, focusing among other things on the optimisation of already applicable detection methods (HPLC-MS) and also on the development of a new innovative quantification method using infusion mass spectrometry.
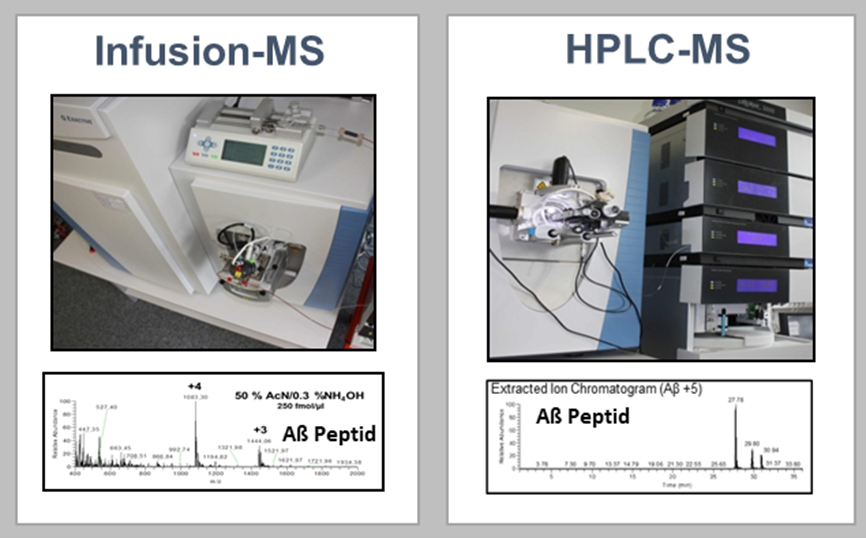
Figure 3: Mass spectrometry-based methods for the detection of amyloid-β peptides.