Medical proteome analysis
Research focus
FUNCTIONAL PROTEOMICS – Neurodegenerative diseases
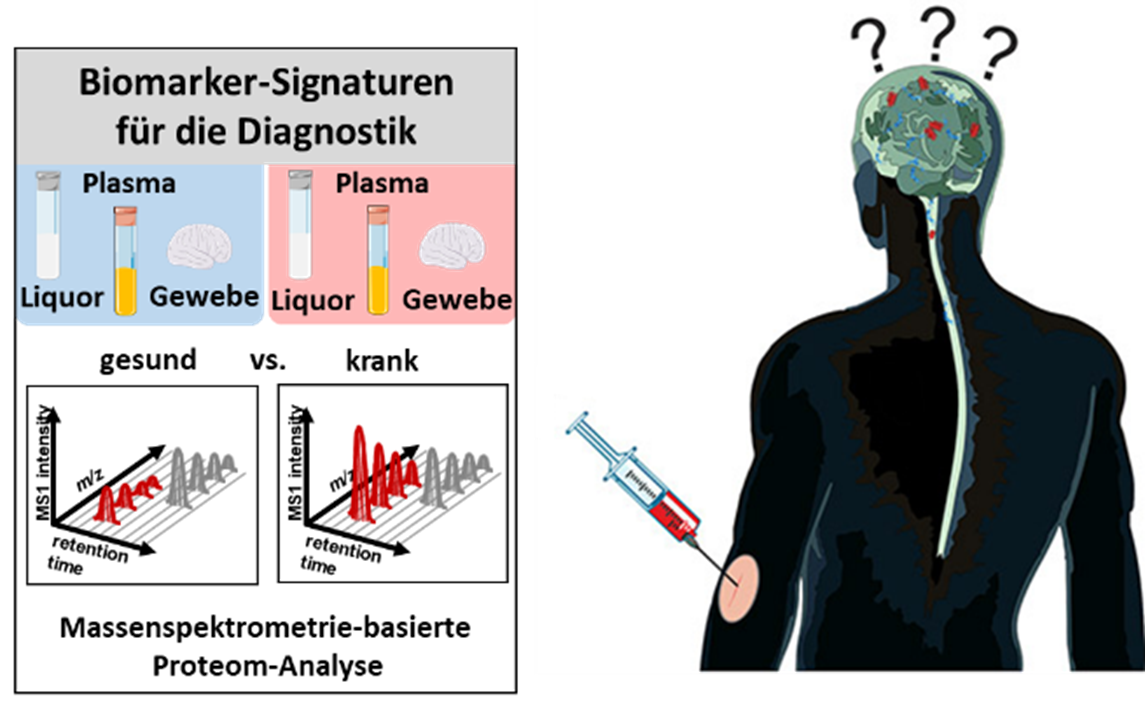
Neurodegenerative diseases are currently not curable. This is due on the one hand to the lack of knowledge of the molecular background of the diseases and on the other hand to the lack of adequate methods for early diagnosis. One focus of the Medical Proteome Analysis competence area is the development of mass spectrometry-based methods for the identification of disease-specific protein signatures for diagnostics and also for a better understanding of the disease processes at the molecular level.
CLINICAL PROTEOMICS – Cancer Diseases
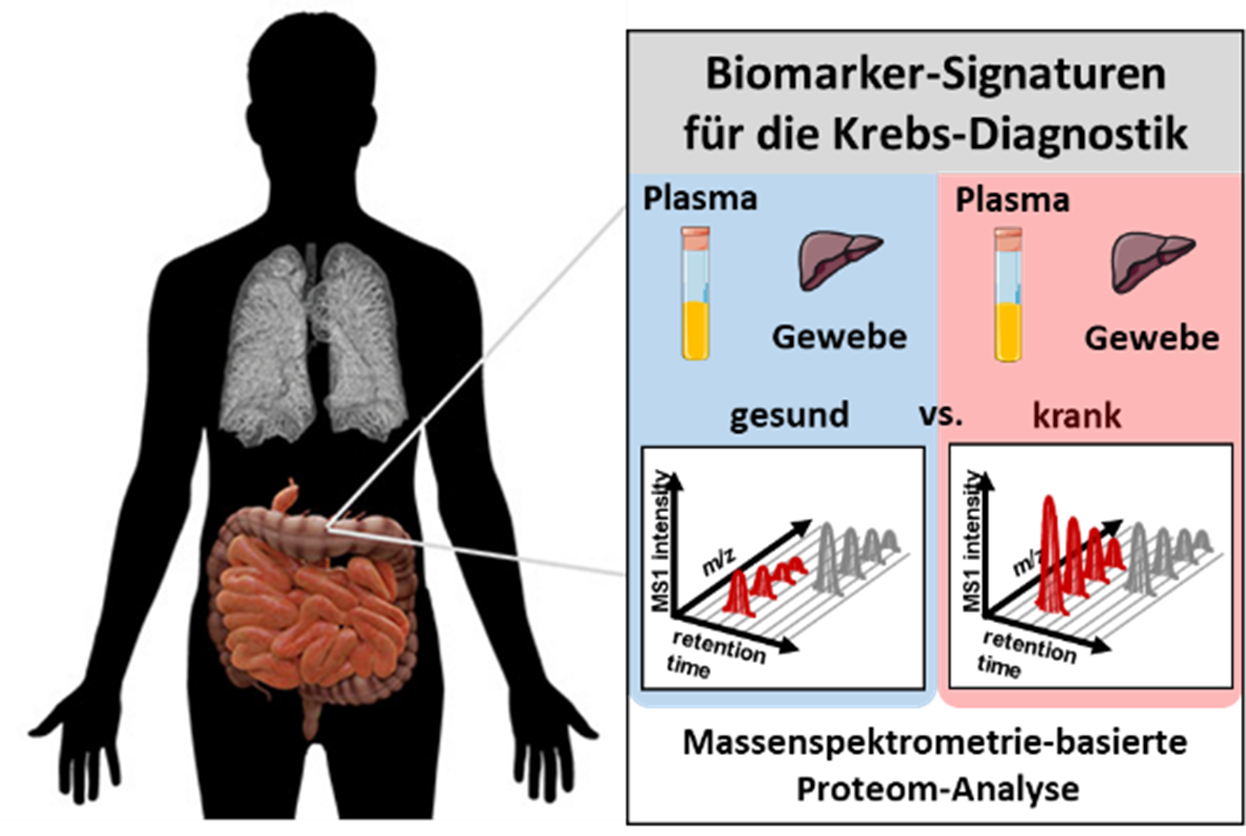
Specific biomarkers are essential for diagnosing cancers earlier and predicting their course in order to improve therapy. One focus of the Medical Proteome Analysis competence area is the identification of protein biomarkers that can be used for differential diagnostics but also as therapeutic target molecules. For this purpose, we examine cell culture models, tissue and body fluids using mass spectrometry-based methods.
Medical Bioinfomatics
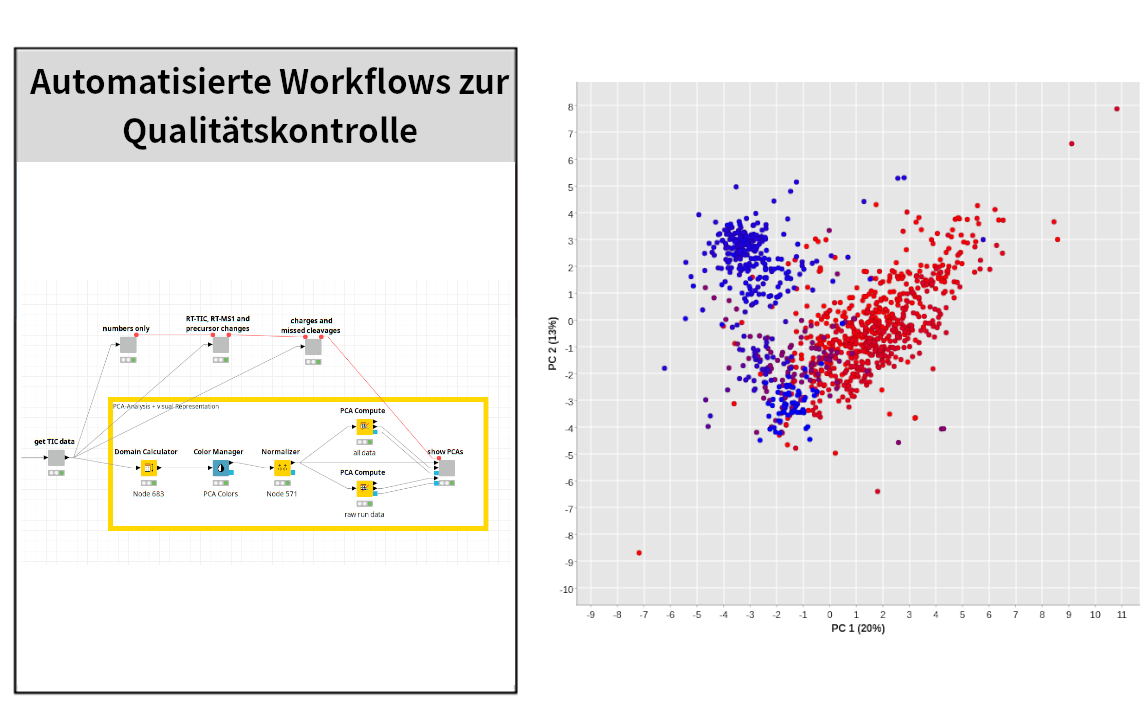
The bioinformatics of proteomics works methodologically closely with the competence area of medical proteome analysis and carries out the bioinformatic and biostatistical evaluations of the measurement data. Here, not only are already established workflows used for the biomarker detection of neurological and oncological diseases, but new bioinformatic methods are also developed and existing evaluation software optimised. For example, methods for the for the inference of proteins from peptide measurements, workflows for bioinformatics workflows for the bioinformatic and biostatistical evaluation of biostatistical analysis of proteome data, as well as the rapid search of all possible possible tryptic peptides.
Method Development
PROTEOMIC METHODS FOR CLINICAL QUESTIONS
Altered proteins play a central role in almost all oncological and neurodegenerative diseases and can therefore provide information about the development and course of the disease and also serve as diagnostic markers. Mass spectrometry-based methods are being developed in the Medical Proteome Analysis competence area.
TISSUE-BASED ANALYSES
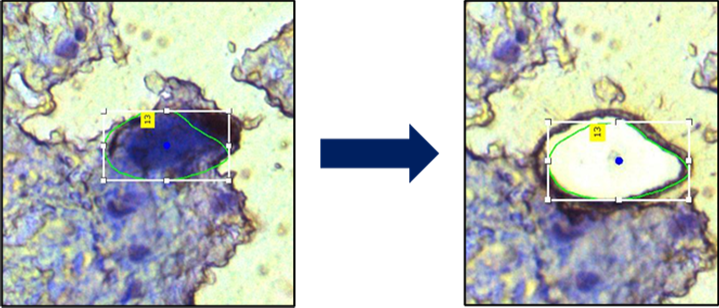
Laser capture microdissection (LCM) or laser microdissection (LMD), enables isolation of defined single cells or cell clusters. In this approach, tissue sections are visualised on slides under the microscope. Cells or regions can then be cut out with the help of a laser and collected in a vessel to finally analyse proteins characteristic for the subpopulation by mass spectrometry.
PROTEIN BIOMARKER DISCOVERY METHODS
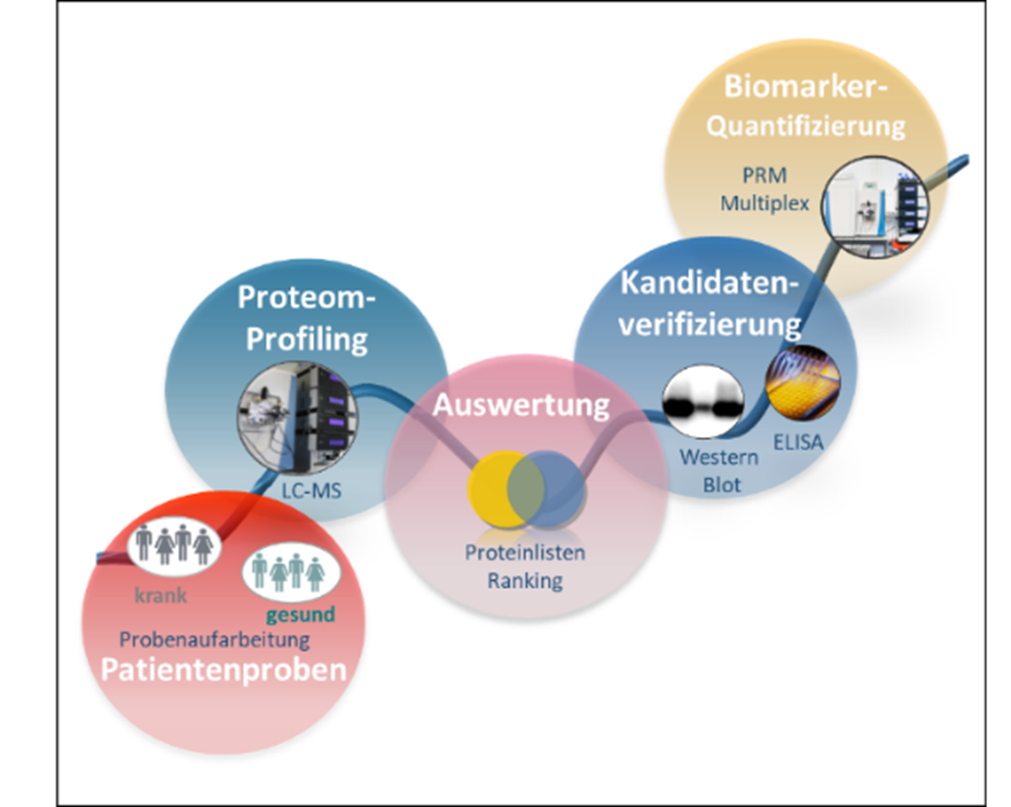
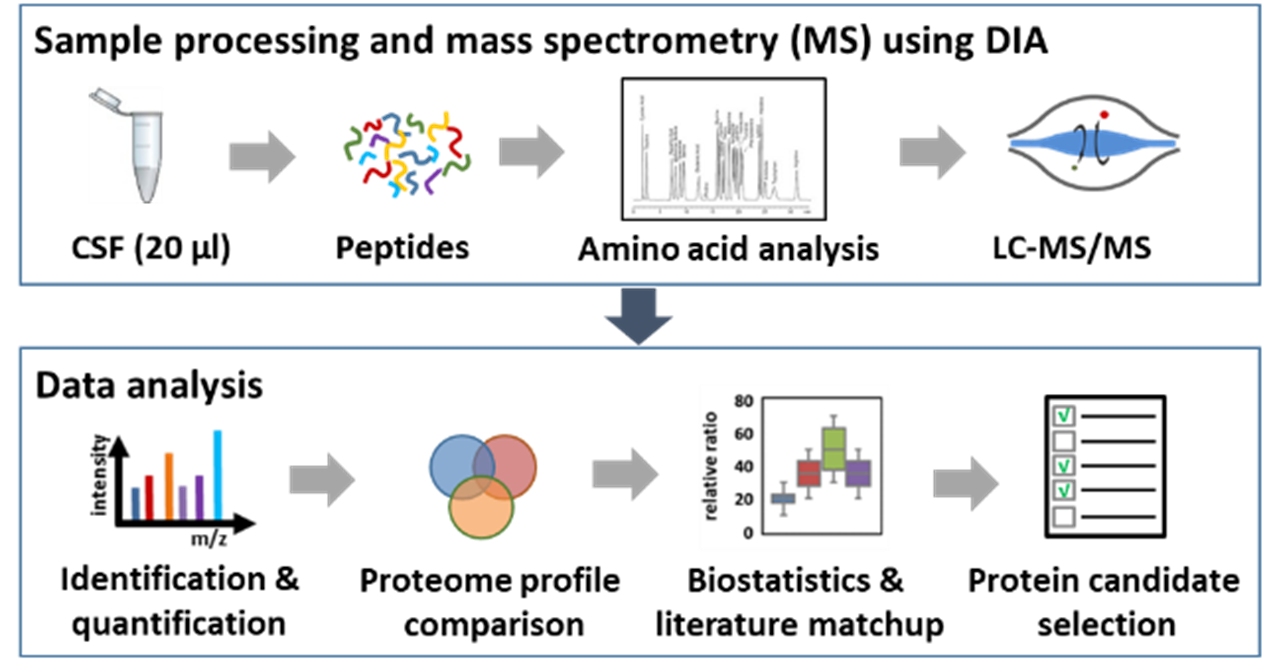
The analysis of body fluids is important for the diagnosis of neurodegenerative or cancer diseases. A molecular measure for diagnostics and thus a possible biomarker for a specific disease is an abnormal protein abundance in body fluids such as cerebrospinal fluid or plasma. However, the quality and processing of the respective body fluid as well as the selection of the appropriate analysis method are key factors for a meaningful result of the analysis. In the competence area of medical proteome analysis, we develop innovative methods and strategies for biomarker identification with a strong focus on establishing applicable standard procedures and defined quality control.
Bioinformatics
The bioinformatics of proteomics works methodologically closely with the competence area of medical proteome analysis and carries out the bioinformatic and biostatistical evaluations of the measurement data. Not only are already established workflows for the biomarker detection of neurological and oncological diseases applied, but new bioinformatic methods are also developed and existing evaluation software optimised.