Imaging techniques
Label-free digital pathology
Tissue and cell diagnostics
One of the most frequently used diagnostic methods is the examination of thin tissue sections. For this purpose, tissue samples are taken and divided into thin sections (1-10 µm) with a microtome. In classical histopathology, which is shown in the upper part of the figure, the sample is stained (H&E) and evaluated under the microscope by a pathologist. The resulting diagnosis is the gold standard in the clinic. However, analogous to tissue samples, clinics also analyse cell samples, which are often obtained from non-invasive or low-invasive body fluids such as urine from patients. In the case of bladder cancer, for example, the gold standard for diagnosis is invasive cystoscopy (tissue diagnostics), which can cause complications. In addition to this method, urinary cytology, i.e. the microscopic evaluation of H&E-stained cells from the urine, is used as an accompanying method. We have established an alternative, label-free approach. We call this approach label-free digital pathology (Kallenbach-Thieltges et al Journal of Biophotonics 2013).
Label-free digital pathology
For this purpose, spatially resolved vibration spectra are recorded using IR/Raman imaging. The vibration spectra represent the chemical composition of the underlying tissue or cells. We examine human tissue or cells with IR-/Raman-imaging. The spectra are used as an integral marker for the biochemical status (proteome, genome, transcriptome, lipidome and metabolome) of the tissue or cells at the time of sampling. The spectra form patterns that are characteristic of the molecular status, much like a fingerprint is characteristic of a human being.
The figure shows the workflow of classical histopathology and the classification of tissue by IR/Raman imaging. In IR/Raman imaging, the samples are not stained, but spatially resolved spectra are recorded. By comparison with a spectral database, a label-free, automated classification of tissue and cells is possible. In comparison to the gold standard in the clinic, histopathology, we achieve a sensitivity and specificity of over 95% for various tissues and cells (Kallenbach-Thieltges et al Journal of Biophotonics 2013).
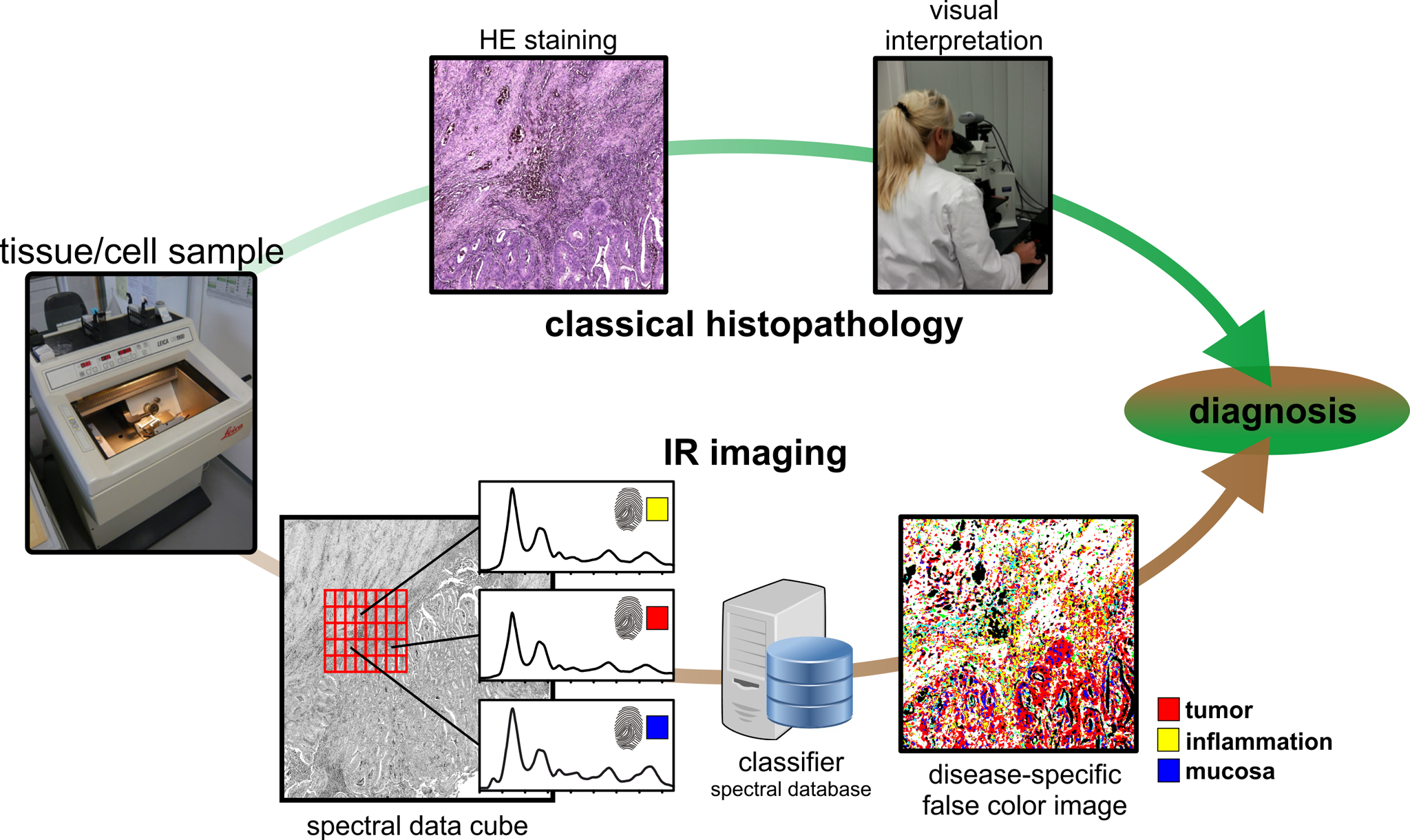
Figure 1: In the upper course the classical histopathology is shown. The tissue is divided into thin sections and stained to make tissue structures visible. The tissue is then assessed by visual inspection. Label-free IR/Raman imaging is shown in the lower part as a further tool. Here the unstained tissue thin section is automatically analysed based on spatially resolved spectra. By means of machine learning, this allows a classification and allocation of the tissue, which is independent of the observer.
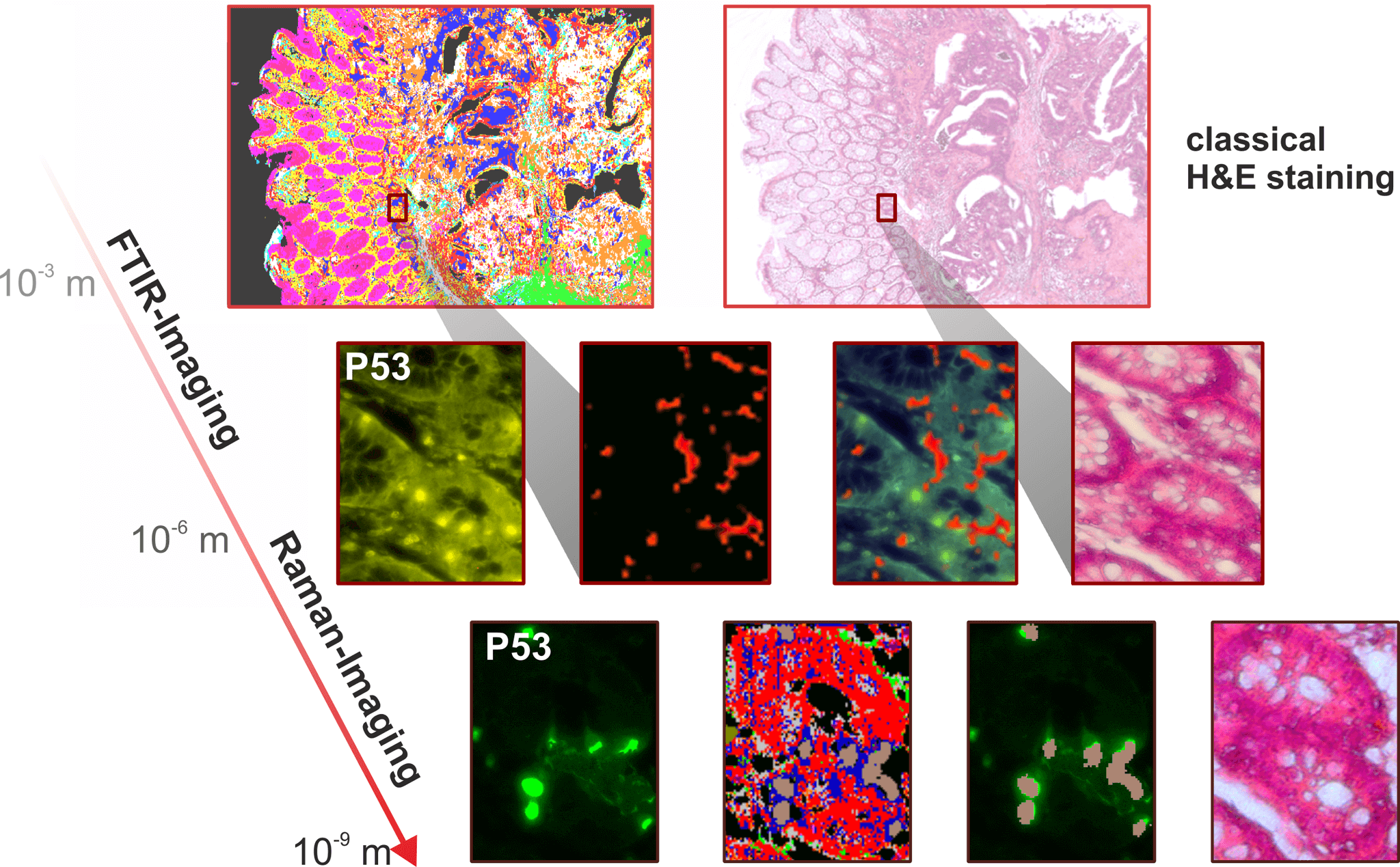
Figure 2: The resolution of a microscope depends on the irradiated wavelength. IR imaging is therefore ideally suited for large area tissue analysis, but when it comes to viewing individual cells, it is necessary to switch to a higher resolution technique, Raman imaging.
Infrared imaging for tissue diagnostics
IR imaging is carried out with infrared microscopes using special FPA (focal plane array) detectors with 64×64 or 128×128 pixels or microbolometers with 480×480 pixels. This enables the simultaneous spatially resolved measurement of large tissue sections. Further technical modifications allow a fast and continuous data acquisition. Classically, the FTIR method (Fourier Transform Infrared) was used, but this is technically very slow for imaging. In the meantime, quantum cascade lasers (QCL) have been established, which allow IR imaging in a few minutes instead of many hours (Kuepper et al Scientific Reports 2018).
Raman imaging for tissue and cell diagnostics
Raman imaging is performed with confocal microscopes in combination with three lasers (532, 633 and 785 nm wavelengths), ultra-high throughput spectrometers and back-thinned, deep depletion CCD detectors. This enables the spatially resolved measurement of samples. However, the conventional Raman technique is slow for imaging. Therefore, nonlinear Raman methods, including coherent anti-Stokes Raman scattering (CARS) and stimulated Raman scattering (SRS), have been established using tunable picosecond lasers. This allows fast imaging at speeds up to video rate instead of several hours (Aljakouch et al Analytical Chemistry 2019, Krauß et al Journal of Biophotonics 2018, Yosef et al Analytical Chemistry 2017, Petersen et al Analyst 2016).
Molecular fingerprints for more precise diagnostics
In addition to medical diagnostic applications, FTIR/Raman imaging can also be used in biomarker searches due to its robust, spatially resolved and observer-independent characterisation on untreated tissue samples or cells. For this purpose we have developed a unique method in ProDi, which uses the spatially resolved tissue recognition of IR-imaging to collect samples for omics techniques (e.g. proteomics) using laser microdissection (LMD). This allows for the first time in the field of biomarker search to work in spatial resolution on very precisely annotated but still untreated samples (Großerüschkamp et al Scientific Reports 2017).
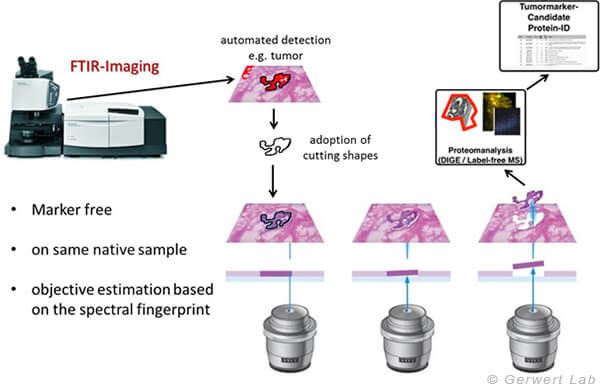
Figure 3: The advantage of the methods described here is that the tissue is not altered and is therefore available unchanged for other methods, such as proteomics. This is a unique advantage in biomarker searches. The method developed in ProDi allows the automated collection of homogeneous tissue areas by combining label-free digital pathology with laser microdissection.
Press
Press releases
Press review
External Funding

111.08.03.05-133974
Validierung und Standardisierung des Workflows zur Proteindiagnostik
FP-0259 CDE
Verifizierung von neuen molekularen Markern für die Frühdiagnose von Lungentumoren zum Nutzen der nachgehenden Vorsorge
Marker-gestützte Nachsorge von Patienten mit nicht-muskelinvasiven low/intermediate-risk Harnblasentumoren (UroFollow)

PURE – Protein Research Unit Ruhr within Europe
233-1.08.03.03-031-68079
Validierung Marker-freier Imaging Verfahren und neu identifizierter Biomarker unter Nutzung des PURE-Konsortium
FP-339
Entwicklung proteinanalytischer Verfahren zur Identifikation von Kandidatenmarkern zur Unterstützung der (Früh-)Diagnose asbestassoziierter Lungen- und Pleuratumoren